Abstract
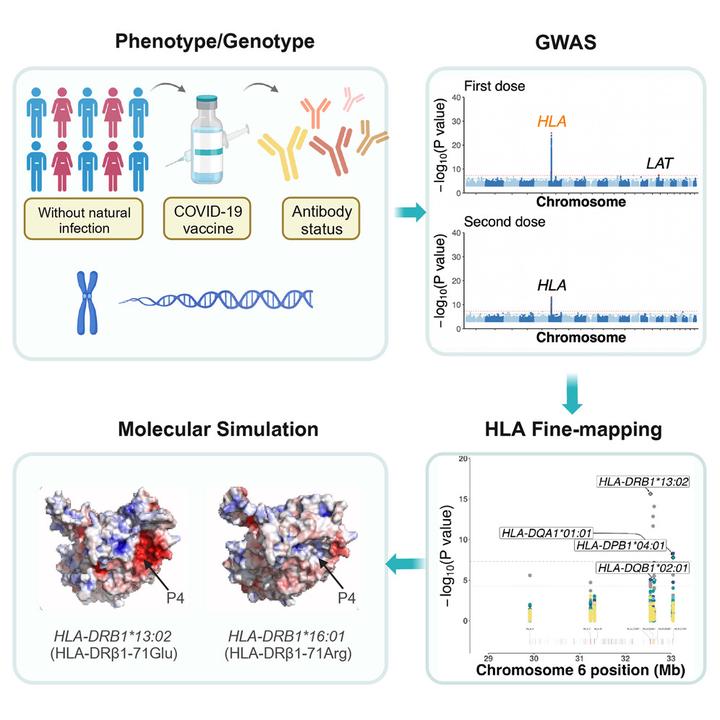
Human humoral immune responses to SARS-CoV-2 vaccines exhibit substantial inter-individual variability and have been linked to vaccine efficacy. To elucidate the underlying mechanism behind this variability, we conducted a genome-wide association study (GWAS) on the anti-spike IgG serostatus of UK Biobank participants who were previously uninfected by SARS-CoV-2 and had received either the first dose (n = 54,066) or the second dose (n = 46,232) of COVID-19 vaccines. Our analysis revealed significant genome-wide associations between the IgG antibody serostatus following the initial vaccine and human leukocyte antigen (HLA) class II alleles. Specifically, the HLA-DRB1∗13:02 allele (MAF = 4.0%, OR = 0.75, p = 2.34e−16) demonstrated the most statistically significant protective effect against IgG seronegativity. This protective effect was driven by an alteration from arginine (Arg) to glutamic acid (Glu) at position 71 on HLA-DRβ1 (p = 1.88e−25), leading to a change in the electrostatic potential of pocket 4 of the peptide binding groove. Notably, the impact of HLA alleles on IgG responses was cell type specific, and we observed a shared genetic predisposition between IgG status and susceptibility/severity of COVID-19. These results were replicated within independent cohorts where IgG serostatus was assayed by two different antibody serology tests. Our findings provide insights into the biological mechanism underlying individual variation in responses to COVID-19 vaccines and highlight the need to consider the influence of constitutive genetics when designing vaccination strategies for optimizing protection and control of infectious disease across diverse populations.